RNA-seq Tam on Acinetobacter baumannii strain ATCC 19606 CP059040.1 (Data_Tam_RNAseq_2024)
gene_x 0 like s 843 view s
Tags: bacterium, pipeline, RNA-seq
http://xgenes.com/article/article-content/209/rna-seq-skin-organoids-on-grch38-chrhsv1-final/ http://xgenes.com/article/article-content/157/prepare-virus-gtf-for-nextflow-run/
Methods
Data was processed using nf-core/rnaseq v3.12.0 (doi: https://doi.org/10.5281/zenodo.1400710) of the nf-core collection of workflows (Ewels et al., 2020).
The pipeline was executed with Nextflow v22.10.5 (Di Tommaso et al., 2017) with the following command:
nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results --fasta /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040.fasta --gff /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040.gff -profile docker -resume --max_cpus 55 --max_memory 512.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner star_salmon --gtf_group_features gene_id --gtf_extra_attributes gene_name --featurecounts_group_type gene_biotype --featurecounts_feature_type transcript
-
Preparing raw data
They are wildtype strains grown in different medium. AUM - artificial urine medium Urine - human urine MHB - Mueller-Hinton broth AUM(人工尿液培养基):pH值、营养成分、无菌性、渗透压、温度、污染物。 Urine(人类尿液):pH值、比重、温度、污染物、化学成分、微生物负荷。 MHB(Mueller-Hinton培养基):pH值、无菌性、营养成分、温度、渗透压、抗生素浓度。 mkdir raw_data; cd raw_data ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-1/AUM-1_1.fq.gz AUM_r1_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-1/AUM-1_2.fq.gz AUM_r1_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-2/AUM-2_1.fq.gz AUM_r2_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-2/AUM-2_2.fq.gz AUM_r2_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-3/AUM-3_1.fq.gz AUM_r3_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/AUM-3/AUM-3_2.fq.gz AUM_r3_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-1/MHB-1_1.fq.gz MHB_r1_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-1/MHB-1_2.fq.gz MHB_r1_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-2/MHB-2_1.fq.gz MHB_r2_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-2/MHB-2_2.fq.gz MHB_r2_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-3/MHB-3_1.fq.gz MHB_r3_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/MHB-3/MHB-3_2.fq.gz MHB_r3_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-1/Urine-1_1.fq.gz Urine_r1_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-1/Urine-1_2.fq.gz Urine_r1_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-2/Urine-2_1.fq.gz Urine_r2_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-2/Urine-2_2.fq.gz Urine_r2_R2.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-3/Urine-3_1.fq.gz Urine_r3_R1.fq.gz ln -s ../X101SC24105589-Z01-J001/01.RawData/Urine-3/Urine-3_2.fq.gz Urine_r3_R2.fq.gz -
(Optional) using trinity to find the most closely reference
Trinity --seqType fq --max_memory 50G --left trimmed/wt_r1_R1.fastq.gz --right trimmed/wt_r1_R2.fastq.gz --CPU 12 #https://www.genome.jp/kegg/tables/br08606.html#prok acb KGB Acinetobacter baumannii ATCC 17978 2007 GenBank abm KGB Acinetobacter baumannii SDF 2008 GenBank aby KGB Acinetobacter baumannii AYE 2008 GenBank abc KGB Acinetobacter baumannii ACICU 2008 GenBank abn KGB Acinetobacter baumannii AB0057 2008 GenBank abb KGB Acinetobacter baumannii AB307-0294 2008 GenBank abx KGB Acinetobacter baumannii 1656-2 2012 GenBank abz KGB Acinetobacter baumannii MDR-ZJ06 2012 GenBank abr KGB Acinetobacter baumannii MDR-TJ 2012 GenBank abd KGB Acinetobacter baumannii TCDC-AB0715 2012 GenBank abh KGB Acinetobacter baumannii TYTH-1 2012 GenBank abad KGB Acinetobacter baumannii D1279779 2013 GenBank abj KGB Acinetobacter baumannii BJAB07104 2013 GenBank abab KGB Acinetobacter baumannii BJAB0715 2013 GenBank abaj KGB Acinetobacter baumannii BJAB0868 2013 GenBank abaz KGB Acinetobacter baumannii ZW85-1 2013 GenBank abk KGB Acinetobacter baumannii AbH12O-A2 2014 GenBank abau KGB Acinetobacter baumannii AB030 2014 GenBank abaa KGB Acinetobacter baumannii AB031 2014 GenBank abw KGB Acinetobacter baumannii AC29 2014 GenBank abal KGB Acinetobacter baumannii LAC-4 2015 GenBank #Note that the Acinetobacter baumannii strain ATCC 19606 chromosome, complete genome (GenBank: CP059040.1) was choosen as reference! -
Downloading CP059040.fasta and CP059040.gff from GenBank
-
(Optional) Preparing CP059040.fasta, CP059040_gene.gff3 and CP059040.bed
#Reference genome: https://www.ncbi.nlm.nih.gov/nuccore/CP059040 cp /media/jhuang/Elements2/Data_Tam_RNASeq3/CP059040.fasta . # Elements (Anna C.arnes) cp /media/jhuang/Elements2/Data_Tam_RNASeq3/CP059040_gene.gff3 . cp /media/jhuang/Elements2/Data_Tam_RNASeq3/CP059040_gene.gtf . cp /media/jhuang/Elements2/Data_Tam_RNASeq3/CP059040.bed . rsync -a -P CP059040.fasta jhuang@hamm:~/DATA/Data_Tam_RNAseq_2024/ rsync -a -P CP059040_gene.gff3 jhuang@hamm:~/DATA/Data_Tam_RNAseq_2024/ rsync -a -P CP059040.bed jhuang@hamm:~/DATA/Data_Tam_RNAseq_2024/ (base) jhuang@WS-2290C:/media/jhuang/Elements2/Data_Tam_RNASeq3$ find . -name "CP059040*" ./CP059040.fasta ./CP059040.bed ./CP059040.gb ./CP059040.gff3 ./CP059040.gff3_backup ./CP059040_full.gb ./CP059040_gene.gff3 ./CP059040_gene.gtf ./CP059040_gene_old.gff3 ./CP059040_rRNA.gff3 ./CP059040_rRNA_v.gff3 # ---- REF: Acinetobacter baumannii ATCC 17978 (DEBUG, gene_name failed) ---- #gffread -E -F -T GCA_000015425.1_ASM1542v1_genomic.gff -o GCA_000015425.1_ASM1542v1_genomic.gtf_ #grep "CDS" GCA_000015425.1_ASM1542v1_genomic.gtf_ > GCA_000015425.1_ASM1542v1_genomic.gtf #sed -i -e "s/\tCDS\t/\texon\t/g" GCA_000015425.1_ASM1542v1_genomic.gtf #gffread -E -F --bed GCA_000015425.1_ASM1542v1_genomic.gtf -o GCA_000015425.1_ASM1542v1_genomic.bed grep "locus_tag" GCA_000015425.1_ASM1542v1_genomic.gtf_ > GCA_000015425.1_ASM1542v1_genomic.gtf sed -i -e "s/\ttranscript\t/\texon\t/g" GCA_000015425.1_ASM1542v1_genomic.gtf # or using fc_count_type=transcript sed -i -e "s/\tgene_name\t/\tName\t/g" GCA_000015425.1_ASM1542v1_genomic.gtf gffread -E -F --bed GCA_000015425.1_ASM1542v1_genomic.gtf -o GCA_000015425.1_ASM1542v1_genomic.bed #grep "gene_name" GCA_000015425.1_ASM1542v1_genomic.gtf | wc -l #69=3887-3803 cp CP059040.gff3 CP059040_backup.gff3 sed -i -e "s/\tGenbank\tgene\t/\tGenbank_gene\t/g" CP059040.gff3 grep "Genbank_gene" CP059040.gff3 > CP059040_gene.gff3 sed -i -e "s/\tGenbank_gene\t/\tGenbank\tgene\t/g" CP059040_gene.gff3 #3796-3754=42--> they are pseudogene since grep "pseudogene" CP059040.gff3 | wc -l = 42 # -------------------------------------------------------------------------------------------------------------------------------------------------- # ---------- PREPARING gff3 file including gene_biotype=protein_coding+gene_biotype=tRNA = total(3754)) and gene_biotype=pseudogene(42) ------------ cp CP059040.gff3 CP059040_backup.gff3 sed -i -e "s/\tGenbank\tgene\t/\tGenbank_gene\t/g" CP059040.gff3 grep "Genbank_gene" CP059040.gff3 > CP059040_gene.gff3 sed -i -e "s/\tGenbank_gene\t/\tGenbank\tgene\t/g" CP059040_gene.gff3 grep "gene_biotype=pseudogene" CP059040.gff3_backup >> CP059040_gene.gff3 #-->3796 #The whole point of the GTF format was to standardise certain aspects that are left open in GFF. Hence, there are many different valid ways to encode the same information in a valid GFF format, and any parser or converter needs to be written specifically for the choices the author of the GFF file made. For example, a GTF file requires the gene ID attribute to be called "gene_id", while in GFF files, it may be "ID", "Gene", something different, or completely missing. # from gff3 to gtf sed -i -e "s/\tID=gene-/\tgene_id \"/g" CP059040_gene.gtf sed -i -e "s/;/\"; /g" CP059040_gene.gtf sed -i -e "s/=/=\"/g" CP059040_gene.gtf #sed -i -e "s/\n/\"\n/g" CP059040_gene.gtf #using editor instead! #The following is GTF-format. CP000521.1 Genbank exon 95 1492 . + . transcript_id "gene0"; gene_id "gene0"; Name "A1S_0001"; gbkey "Gene"; gene_biotype "protein_coding"; locus_tag "A1S_0001"; #NZ_MJHA01000001.1 RefSeq region 1 8663 . + . ID=id0;Dbxref=taxon:575584;Name=unnamed1;collected-by=IG Schaub;collection-date=1948;country=USA: Vancouver;culture-collection=ATCC:19606;gbkey=Src;genome=plasmid;isolation-source=urine;lat-lon=37.53 N 75.4 W;map=unlocalized;mol_type=genomic DNA;nat-host=Homo sapiens;plasmid-name=unnamed1;strain=ATCC 19606;type-material=type strain of Acinetobacter baumannii #NZ_MJHA01000001.1 RefSeq gene 228 746 . - . ID=gene0;Name=BIT33_RS00005;gbkey=Gene;gene_biotype=protein_coding;locus_tag=BIT33_RS00005;old_locus_tag=BIT33_18795 #NZ_MJHA01000001.1 Protein Homology CDS 228 746 . - 0 ID=cds0;Parent=gene0;Dbxref=Genbank:WP_000839337.1;Name=WP_000839337.1;gbkey=CDS;inference=COORDINATES: similar to AA sequence:RefSeq:WP_000839337.1;product=hypothetical protein;protein_id=WP_000839337.1;transl_table=11 ##gff-version 3 ##sequence-region CP059040.1 1 3980852 ##species https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=470 gffread -E -F --bed CP059040.gff3 -o CP059040.bed #-->3796 ##prepare the GTF-format (see above) --> ERROR! ----> using CP059040.gff3 ##stringtie adeIJ.abx_r1.sorted.bam -o adeIJ.abx_r1.sorted_transcripts.gtf -v -G /media/jhuang/Elements/Data_Tam_RNASeq3/CP059040.gff3 -A adeIJ.abx_r1.sorted.gene_abund.txt -C adeIJ.abx_r1.sorted.bam.cov_refs.gtf -e -b adeIJ.abx_r1.sorted_ballgown #[01/21 10:57:46] Loading reference annotation (guides).. #GFF warning: merging adjacent/overlapping segments of gene-H0N29_00815 on CP059040.1 (179715-179786, 179788-180810) #[01/21 10:57:46] 3796 reference transcripts loaded. #Default stack size for threads: 8388608 #WARNING: no reference transcripts found for genomic sequence "gi|1906906720|gb|CP059040.1|"! (mismatched reference names?) #WARNING: no reference transcripts were found for the genomic sequences where reads were mapped! #Please make sure the -G annotation file uses the same naming convention for the genome sequences. #[01/21 10:58:30] All threads finished. # ERROR: failed to find the gene identifier attribute in the 9th column of the provided GTF file. # The specified gene identifier attribute is 'Name' # An example of attributes included in your GTF annotation is 'ID=exon-H0N29_00075-1;Parent=rna-H0N29_00075;gbkey=rRNA;locus_tag=H0N29_00075;product=16S ribosomal RNA' # The program has to termin # ERROR: failed to find the gene identifier attribute in the 9th column of the provided GTF file. # The specified gene identifier attribute is 'gene_biotype' # An example of attributes included in your GTF annotation is 'ID=exon-H0N29_00075-1;Parent=rna-H0N29_00075;gbkey=rRNA;locus_tag=H0N29_00075;product=16S ribosomal RNA' # The program has to terminate. #grep "ID=cds-" CP059040.gff3 | wc -l #grep "ID=exon-" CP059040.gff3 | wc -l #grep "ID=gene-" CP059040.gff3 | wc -l #the same as H0N29_18980/5=3796 grep "gbkey=" CP059040.gff3 | wc -l 7695 grep "ID=id-" CP059040.gff3 | wc -l 5 grep "locus_tag=" CP059040.gff3 | wc -l 7689 #... cds 3701 locus_tag=xxxx, no gene_biotype exon 96 locus_tag=xxxx, no gene_biotype gene 3796 locus_tag=xxxx, gene_biotype=xxxx, id (riboswitch+direct_repeat,5) both no --> ignoring them!! # grep "ID=id-" CP059040.gff3 rna 96 locus_tag=xxxx, no gene_biotype ------------------ 7694 cp CP059040.gff3_backup CP059040.gff3 grep "^##" CP059040.gff3 > CP059040_gene.gff3 grep "ID=gene" CP059040.gff3 >> CP059040_gene.gff3 #!!!!VERY_IMPORTANT!!!!: change type '\tCDS\t' to '\texon\t'! sed -i -e "s/\tgene\t/\texon\t/g" CP059040_gene.gff3 -
Preparing the directory trimmed
mkdir trimmed trimmed_unpaired; for sample_id in AUM_r1 AUM_r2 AUM_r3 Urine_r1 Urine_r2 Urine_r3 MHB_r1 MHB_r2 MHB_r3; do \ for sample_id in MHB_r1 MHB_r2 MHB_r3; do \ java -jar /home/jhuang/Tools/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads 100 raw_data/${sample_id}_R1.fq.gz raw_data/${sample_id}_R2.fq.gz trimmed/${sample_id}_R1.fq.gz trimmed_unpaired/${sample_id}_R1.fq.gz trimmed/${sample_id}_R2.fq.gz trimmed_unpaired/${sample_id}_R2.fq.gz ILLUMINACLIP:/home/jhuang/Tools/Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:TRUE LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 AVGQUAL:20; done 2> trimmomatic_pe.log; done -
Preparing samplesheet.csv
sample,fastq_1,fastq_2,strandedness AUM_r1,AUM_r1_R1.fq.gz,AUM_r1_R2.fq.gz,auto AUM_r2,AUM_r2_R1.fq.gz,AUM_r2_R2.fq.gz,auto AUM_r3,AUM_r3_R1.fq.gz,AUM_r3_R2.fq.gz,auto MHB_r1,MHB_r1_R1.fq.gz,MHB_r1_R2.fq.gz,auto MHB_r2,MHB_r2_R1.fq.gz,MHB_r2_R2.fq.gz,auto MHB_r3,MHB_r3_R1.fq.gz,MHB_r3_R2.fq.gz,auto Urine_r1,Urine_r1_R1.fq.gz,Urine_r1_R2.fq.gz,auto Urine_r2,Urine_r2_R1.fq.gz,Urine_r2_R2.fq.gz,auto Urine_r3,Urine_r3_R1.fq.gz,Urine_r3_R2.fq.gz,auto -
nextflow run
#Example1: http://xgenes.com/article/article-content/157/prepare-virus-gtf-for-nextflow-run/ docker pull nfcore/rnaseq ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq #Default: --gtf_group_features 'gene_id' --gtf_extra_attributes 'gene_name' --featurecounts_group_type 'gene_biotype' --featurecounts_feature_type 'exon' #(host_env) !NOT_WORKING! jhuang@WS-2290C:~/DATA/Data_Tam_RNAseq_2024$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results --fasta "/home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040.fasta" --gff "/home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040.gff" -profile docker -resume --max_cpus 55 --max_memory 512.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner 'star_salmon' --gtf_group_features 'gene_id' --gtf_extra_attributes 'gene_name' --featurecounts_group_type 'gene_biotype' --featurecounts_feature_type 'transcript' # -- DEBUG_1 (CDS --> exon in CP059040.gff) -- #Checking the record (see below) in results/genome/CP059040.gtf #In ./results/genome/CP059040.gtf e.g. "CP059040.1 Genbank transcript 1 1398 . + . transcript_id "gene-H0N29_00005"; gene_id "gene-H0N29_00005"; gene_name "dnaA"; Name "dnaA"; gbkey "Gene"; gene "dnaA"; gene_biotype "protein_coding"; locus_tag "H0N29_00005";" #--featurecounts_feature_type 'transcript' returns only the tRNA results #Since the tRNA records have "transcript and exon". In gene records, we have "transcript and CDS". replace the CDS with exon grep -P "\texon\t" CP059040.gff | sort | wc -l #96 grep -P "cmsearch\texon\t" CP059040.gff | wc -l #=10 ignal recognition particle sRNA small typ, transfer-messenger RNA, 5S ribosomal RNA grep -P "Genbank\texon\t" CP059040.gff | wc -l #=12 16S and 23S ribosomal RNA grep -P "tRNAscan-SE\texon\t" CP059040.gff | wc -l #tRNA 74 wc -l star_salmon/AUM_r3/quant.genes.sf #--featurecounts_feature_type 'transcript' results in 96 records! grep -P "\tCDS\t" CP059040.gff | wc -l #3701 sed 's/\tCDS\t/\texon\t/g' CP059040.gff > CP059040_m.gff grep -P "\texon\t" CP059040_m.gff | sort | wc -l #3797 # -- DEBUG_2: combination of 'CP059040_m.gff' and 'exon' results in ERROR, using 'transcript' instead! --gff "/home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_m.gff" --featurecounts_feature_type 'transcript' # ---- SUCCESSFUL with directly downloaded gff3 and fasta from NCBI using docker after replacing 'CDS' with 'exon' ---- (host_env) /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results --fasta "/home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040.fasta" --gff "/home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_m.gff" -profile docker -resume --max_cpus 55 --max_memory 512.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner 'star_salmon' --gtf_group_features 'gene_id' --gtf_extra_attributes 'gene_name' --featurecounts_group_type 'gene_biotype' --featurecounts_feature_type 'transcript' # -- DEBUG_3: make sure the header of fasta is the same to the *_m.gff file -
Import data and pca-plot
#mamba activate r_env #install.packages("ggfun") # Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) #library("org.Hs.eg.db") library(dplyr) library(tidyverse) #install.packages("devtools") #devtools::install_version("gtable", version = "0.3.0") library(gplots) library("RColorBrewer") #install.packages("ggrepel") library("ggrepel") # install.packages("openxlsx") library(openxlsx) library(EnhancedVolcano) library(DESeq2) setwd("~/DATA/Data_Tam_RNAseq_2024/results/star_salmon") # Define paths to your Salmon output quantification files files <- c("AUM_r1" = "./AUM_r1/quant.sf", "AUM_r2" = "./AUM_r2/quant.sf", "AUM_r3" = "./AUM_r3/quant.sf", "Urine_r1" = "./Urine_r1/quant.sf", "Urine_r2" = "./Urine_r2/quant.sf", "Urine_r3" = "./Urine_r3/quant.sf", "MHB_r1" = "./MHB_r1/quant.sf", "MHB_r2" = "./MHB_r2/quant.sf", "MHB_r3" = "./MHB_r3/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples replicate <- factor(c("r1", "r2", "r3", "r1", "r2", "r3", "r1", "r2", "r3")) condition <- factor(c("AUM","AUM","AUM", "Urine","Urine","Urine", "MHB","MHB","MHB")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) # -- transcript-level count data (x2) -- # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) write.csv(counts(dds), file="transcript_counts.csv") # -- gene-level count data (x2) -- # Read in the tx2gene map from salmon_tx2gene.tsv tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition+replicate) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #3796->3487 write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) #We don't need to run DESeq(dds) before estimateSizeFactors(dds). In fact, the typical workflow in DESeq2 is the opposite: we usually run estimateSizeFactors(dds) (and other preprocessing functions) before running the main DESeq(dds) function. #The estimateSizeFactors function is used to calculate size factors for normalization, which corrects for differences in library size (i.e., the number of read counts) between samples. This normalization step is crucial to ensure that differences in gene expression aren't merely due to differences in sequencing depth between samples. #The DESeq function, on the other hand, performs the main differential expression analysis, comparing gene expression between different conditions or groups. #So, the typical workflow is: # - Create the DESeqDataSet object. # - Use estimateSizeFactors to normalize for library size. # - (Optionally, estimate dispersion with estimateDispersions if not using the full DESeq function later.) # - Use DESeq for the differential expression analysis. # - However, it's worth noting that if you run the main DESeq function directly after creating the DESeqDataSet object, it will automatically perform the normalization (using estimateSizeFactors) and dispersion estimation steps for you. In that case, there's no need to run estimateSizeFactors separately before DESeq. # draw simple pca and heatmap #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
(Optional (ERROR-->need to be debugged!) ) estimate size factors and dispersion values.
#Size Factors: These are used to normalize the read counts across different samples. The size factor for a sample accounts for differences in sequencing depth (i.e., the total number of reads) and other technical biases between samples. After normalization with size factors, the counts should be comparable across samples. Size factors are usually calculated in a way that they reflect the median or mean ratio of gene expression levels between samples, assuming that most genes are not differentially expressed. #Dispersion: This refers to the variability or spread of gene expression measurements. In RNA-seq data analysis, each gene has its own dispersion value, which reflects how much the counts for that gene vary between different samples, more than what would be expected just due to the Poisson variation inherent in counting. Dispersion is important for accurately modeling the data and for detecting differentially expressed genes. #So in summary, size factors are specific to samples (used to make counts comparable across samples), and dispersion values are specific to genes (reflecting variability in gene expression). sizeFactors(dds) #NULL # Estimate size factors dds <- estimateSizeFactors(dds) # Estimate dispersions dds <- estimateDispersions(dds) #> sizeFactors(dds) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #2.3282468 2.0251928 1.8036883 1.3767551 0.9341929 1.0911693 0.5454526 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.4604461 0.5799834 0.6803681 # (DEBUG) If avgTxLength is Necessary #To simplify the computation and ensure sizeFactors are calculated: assays(dds)$avgTxLength <- NULL dds <- estimateSizeFactors(dds) sizeFactors(dds) #If you want to retain avgTxLength but suspect it is causing issues, you can explicitly instruct DESeq2 to compute size factors without correcting for library size with average transcript lengths: dds <- estimateSizeFactors(dds, controlGenes = NULL, use = FALSE) sizeFactors(dds) # If alone with virus data, the following BUG occured: #Still NULL --> BUG --> using manual calculation method for sizeFactor calculation! HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 data.frame(genes = rownames(dds), dispersions = dispersions(dds)) #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 raw_counts <- counts(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) -
Select the differentially expressed genes
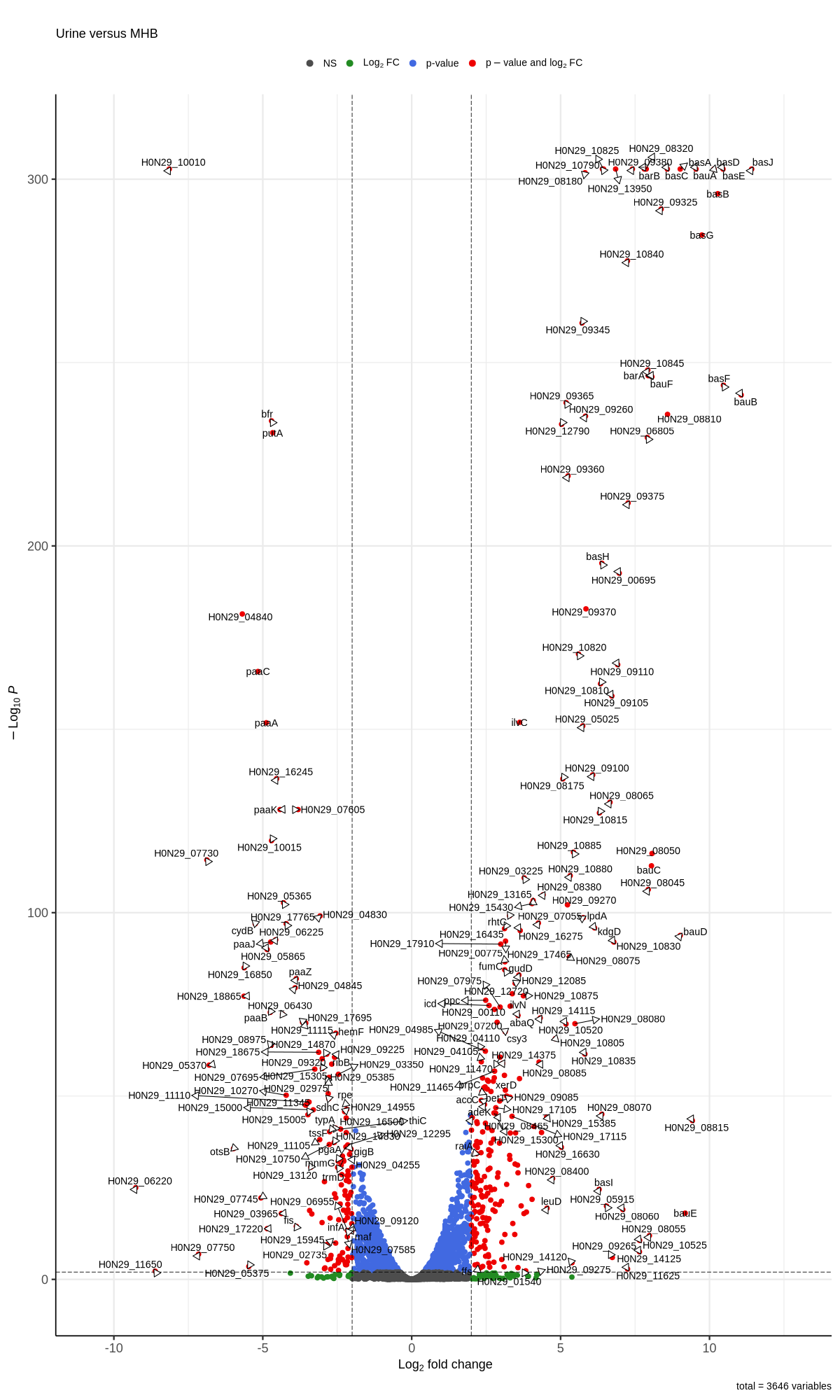
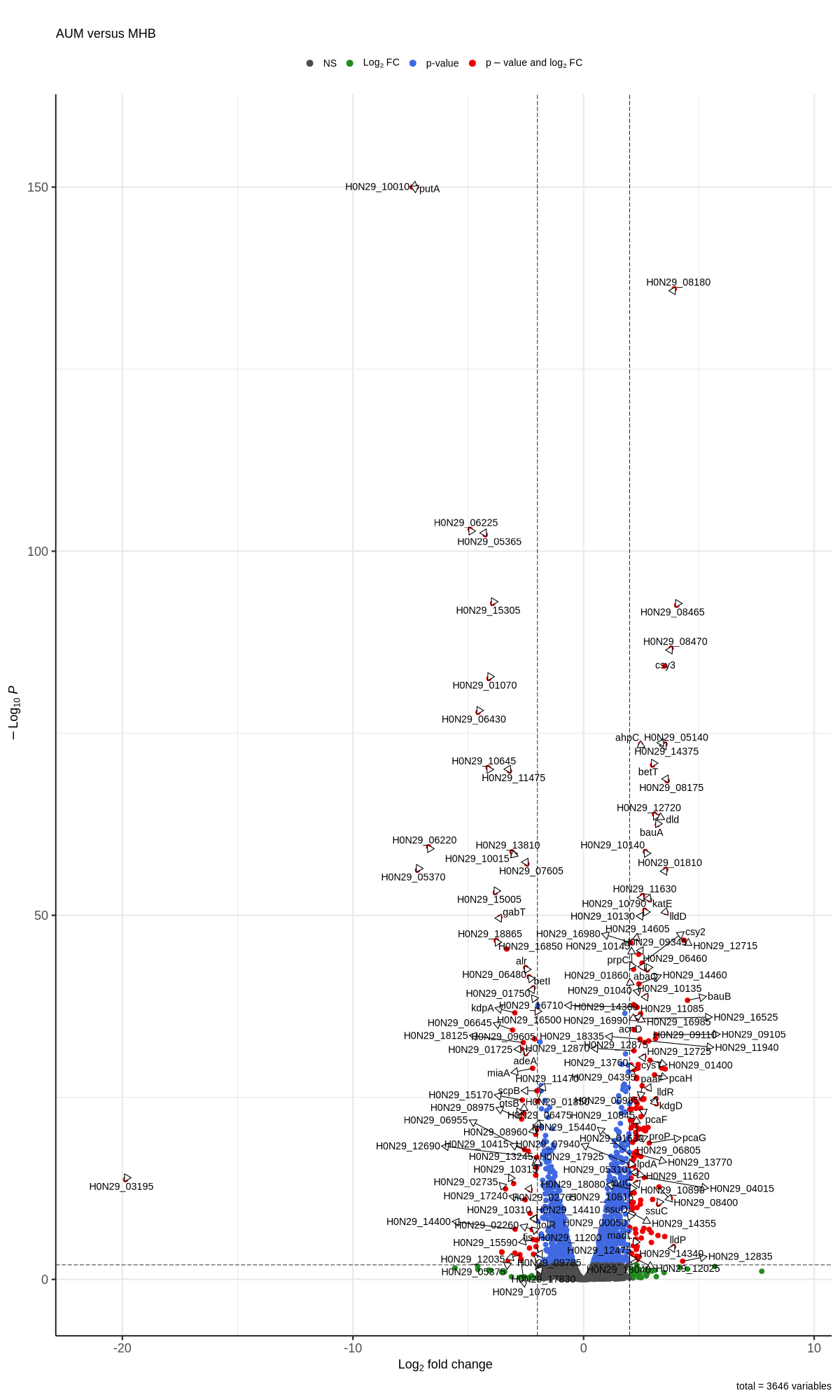
#https://galaxyproject.eu/posts/2020/08/22/three-steps-to-galaxify-your-tool/ #https://www.biostars.org/p/282295/ #https://www.biostars.org/p/335751/ #> dds$condition #[1] AUM AUM AUM Urine Urine Urine MHB MHB MHB #Levels: AUM MHB Urine #CONSOLE: mkdir star_salmon/degenes setwd("degenes") #---- relevel to control ---- dds$condition <- relevel(dds$condition, "MHB") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("AUM_vs_MHB","Urine_vs_MHB") for (i in clist) { contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=1.35) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-1.35) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) } # -- Under host-env -- grep -P "\tgene\t" CP059040.gff > CP059040_gene.gff python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff AUM_vs_MHB-all.txt AUM_vs_MHB-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff AUM_vs_MHB-up.txt AUM_vs_MHB-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff AUM_vs_MHB-down.txt AUM_vs_MHB-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff Urine_vs_MHB-all.txt Urine_vs_MHB-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff Urine_vs_MHB-up.txt Urine_vs_MHB-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024/CP059040_gene.gff Urine_vs_MHB-down.txt Urine_vs_MHB-down.csv #for i in AUM_vs_MHB Urine_vs_MHB; do # echo "contrast = paste(\"condition\", \"${i}\", sep=\"_\")" # echo "res = results(dds, name=contrast)" # echo "res <- res[!is.na(res$log2FoldChange),]" # echo "res_df <- as.data.frame(res)" # #selectLab = selectLab_italics, # echo "png(\"${i}.png\",width=1200, height=1000)" # #legendPosition = 'right',legendLabSize = 12, arrowheads = FALSE, # #subtitle=expression(~Delta*\"$(echo $i | cut -d'_' -f1) versus \" *~Delta*\"$(echo $i | cut -d'_' -f3)\"))" # echo "EnhancedVolcano(res, lab = rownames(res),x = 'log2FoldChange',y = 'padj', pCutoff=5e-2, FCcutoff=1.2, title='', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha=1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle=expression(\"$(echo $i | cut -d'_' -f1) versus $(echo $i | cut -d'_' -f3)\"))" # echo "dev.off()" #done res <- read.csv("AUM_vs_MHB-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] #print(duplicated_genes) # [1] "bfr" "lipA" "ahpF" "pcaF" "alr" "pcaD" "cydB" "lpdA" "pgaC" "ppk1" #[11] "pcaF" "tuf" "galE" "murI" "yccS" "rrf" "rrf" "arsB" "ptsP" "umuD" #[21] "map" "pgaB" "rrf" "rrf" "rrf" "pgaD" "uraH" "benE" #res[res$GeneName == "bfr", ] #1st_strategy First occurrence is kept and Subsequent duplicates are removed #res <- res[!duplicated(res$GeneName), ] #2nd_strategy keep the row with the smallest padj value for each GeneName res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_AUM_vs_MHB.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-150 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("AUM_vs_MHB.png", width=1200, height=2000) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("AUM versus MHB")) dev.off() res <- read.csv("Urine_vs_MHB-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Urine_vs_MHB.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-305 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Urine_vs_MHB.png", width=1200, height=2000) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Urine versus MHB")) dev.off() -
Report
Attached are the results of the analysis. In the Urine_vs_MHB comparison, we identified a total of 259 up-regulated genes (log2FoldChange > 2 and padj < 1e-2) and 138 down-regulated genes (log2FoldChange < -2 and padj < 1e-2) (please refer to the attached volcano plot and Excel files). Notably, the following genes have a p-adjusted value of 0, indicating very high confidence in their differential expression. The bas-series genes (basA, basB, basC, basD, basE, basJ) are particularly prominent: GeneName GeneID baseMean log2FoldChange lfcSE stat pvalue padj basJ gene-H0N29_05120 11166.90 11.42 0.30 38.27 0 0 basE gene-H0N29_05085 12006.52 10.45 0.23 45.76 0 0 basD gene-H0N29_05080 12217.80 10.15 0.24 42.42 0 0 bauA gene-H0N29_05070 25280.68 9.55 0.19 51.48 0 0 basA gene-H0N29_05040 9750.68 9.02 0.18 48.90 0 0 basC gene-H0N29_05075 5034.14 8.58 0.21 40.07 0 0 H0N29_08320 gene-H0N29_08320 4935.78 7.87 0.20 40.01 0 0 barB gene-H0N29_05105 5187.29 7.81 0.18 43.39 0 0 H0N29_09380 gene-H0N29_09380 3477.26 7.41 0.19 38.91 0 0 H0N29_13950 gene-H0N29_13950 13959.05 6.85 0.15 45.70 0 0 H0N29_10825 gene-H0N29_10825 3664.70 6.44 0.17 37.59 0 0 H0N29_10790 gene-H0N29_10790 2574.12 6.41 0.17 37.86 0 0 H0N29_10010 gene-H0N29_10010 9376.84 -8.14 0.19 -43.70 0 0 In the AUM_vs_MHB comparison, we identified a total of 149 up-regulated genes (log2FoldChange > 2 and padj < 1e-2) and 65 down-regulated genes (log2FoldChange < -2 and padj < 1e-2) (please refer to the attached volcano plot and Excel files). The following genes also show a p-adjusted value of 0, indicating very high confidence in their differential expression: GeneName GeneID baseMean log2FoldChange lfcSE stat pvalue padj putA gene-H0N29_09870 36100.24 -7.25 0.15 -49.78 0 0 H0N29_10010 gene-H0N29_10010 9376.84 -7.43 0.18 -41.96 0 0 To ensure proper visualization, I replaced the padj = 0 values with small numbers: 1e-305 for Urine_vs_MHB and 1e-150 for AUM_vs_MHB. We have now identified the significantly expressed genes. If you would like any further analysis based on these genes or need additional plots, please let me know. -
(TODO) clustering the genes and draw heatmap
for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "cut -d',' -f1-1 ${i}-up_annotated.txt > ${i}-up.id"; echo "cut -d',' -f1-1 ${i}-down_annotated.txt > ${i}-down.id"; done cat *.id | sort -u > ids #add Gene_Id in the first line, delete the "" GOI <- read.csv("ids")$Gene_Id #4647 RNASeq.NoCellLine <- assay(rld) #install.packages("gplots") library("gplots") #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman datamat = RNASeq.NoCellLine[GOI, ] #datamat = RNASeq.NoCellLine write.csv(as.data.frame(datamat), file ="gene_expressions.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.05) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=900, height=800) #cex.lab=10, labRow="", png("DEGs_heatmap.png", width=800, height=1000) heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',labRow="", scale='row',trace='none',col=bluered(75), cexCol=1.8, RowSideColors = mycol, margins=c(10,2), cexRow=1.5, srtCol=30, lhei = c(1, 8), lwid=c(2, 8)) #rownames(datamat) #heatmap.2(datamat, Rowv=as.dendrogram(hr), col=bluered(75), scale="row", RowSideColors=mycol, trace="none", margin=c(5,5), sepwidth=c(0,0), dendrogram = 'row', Colv = 'false', density.info='none', labRow="", srtCol=30, lhei=c(0.1,2)) dev.off() #### cluster members ##### write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt') write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt') write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt') write.csv(names(subset(mycl, mycl == '4')),file='cluster4.txt') #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o DEGs_heatmap_cluster_members.xls ~/Tools/csv2xls-0.4/csv_to_xls.py \ significant_gene_expressions.txt \ -d',' -o DEGs_heatmap_expression_data.xls; #### cluster members (advanced) ##### subset_1<-names(subset(mycl, mycl == '1')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_1, ]) #2575 subset_2<-names(subset(mycl, mycl == '2')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_2, ]) #1855 subset_3<-names(subset(mycl, mycl == '3')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_3, ]) #217 subset_4<-names(subset(mycl, mycl == '4')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_4, ]) # subset_5<-names(subset(mycl, mycl == '5')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_5, ]) # # Initialize an empty data frame for the annotated data annotated_data <- data.frame() # Determine total number of genes total_genes <- length(rownames(data)) # Loop through each gene to annotate for (i in 1:total_genes) { gene <- rownames(data)[i] result <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = gene, mart = ensembl) # If multiple rows are returned, take the first one if (nrow(result) > 1) { result <- result[1, ] } # Check if the result is empty if (nrow(result) == 0) { result <- data.frame(ensembl_gene_id = gene, external_gene_name = NA, gene_biotype = NA, entrezgene_id = NA, chromosome_name = NA, start_position = NA, end_position = NA, strand = NA, description = NA) } # Transpose expression values expression_values <- t(data.frame(t(data[gene, ]))) colnames(expression_values) <- colnames(data) # Combine gene information and expression data combined_result <- cbind(result, expression_values) # Append to the final dataframe annotated_data <- rbind(annotated_data, combined_result) # Print progress every 100 genes if (i %% 100 == 0) { cat(sprintf("Processed gene %d out of %d\n", i, total_genes)) } } # Save the annotated data to a new CSV file #write.csv(annotated_data, "cluster1_YELLOW.csv", row.names=FALSE) write.csv(annotated_data, "cluster2_DARKBLUE.csv", row.names=FALSE) write.csv(annotated_data, "cluster3_DARKORANGE.csv", row.names=FALSE) write.csv(annotated_data, "cluster4_DARKMAGENTA.csv", row.names=FALSE) write.csv(annotated_data, "cluster5_DARKCYAN.csv", row.names=FALSE) #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.csv -d',' -o DEGs_heatmap_clusters.xls
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Kraken2 Installation and Usage Guide
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Deciphering S. aureus with metatranscriptomics (Marc's project)
Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
How to correlate RNA-seq Data with Mass Spectrometry Proteomics Data?